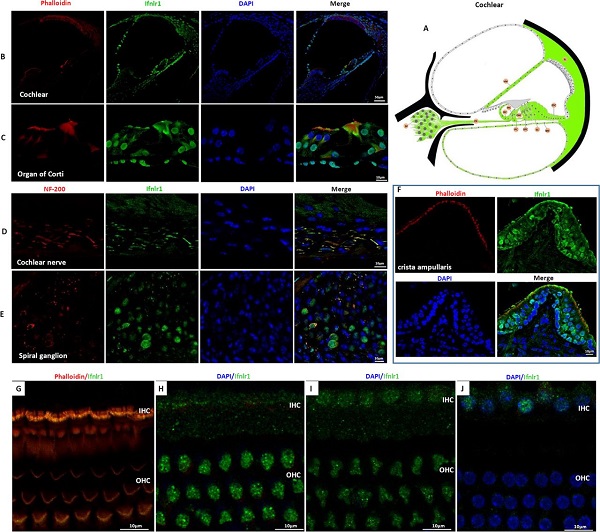
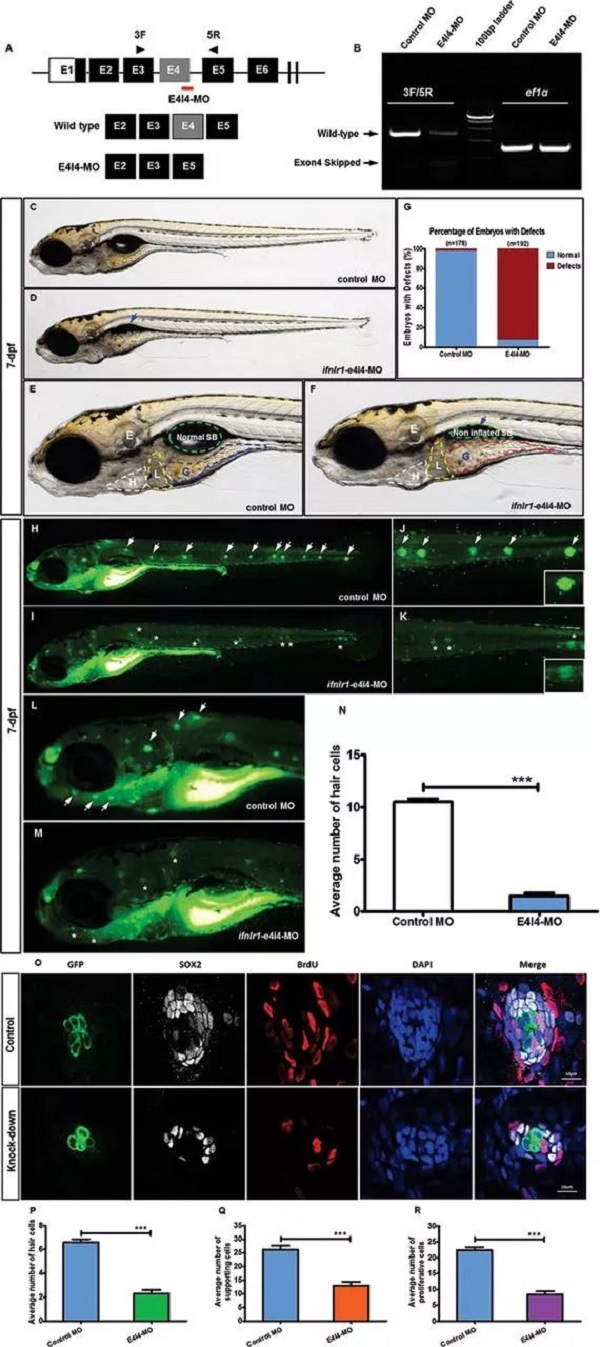
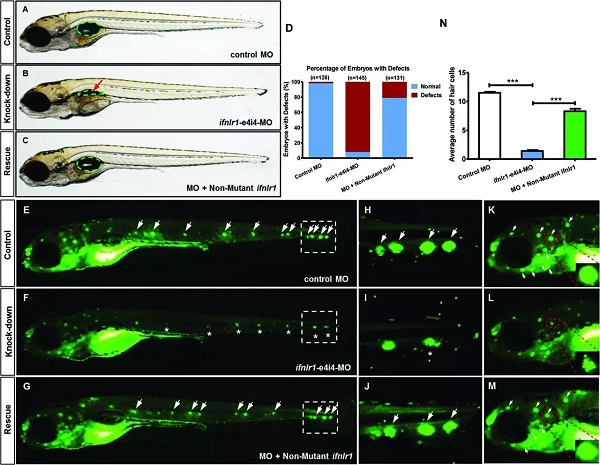
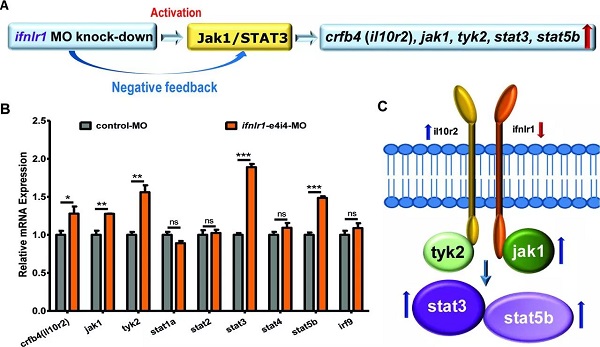
On February 16 (the first day of the first month), the Journal of Medical Genetics published the Journal of Medical Genetics of the People's Liberation Army General Hospital. "Mutation of IFNLR1, an interferon lambda receptor 1, is associated with autosomal-dominant non-syndromic hearing loss ", reveals the important role of IFNLR1 in the auditory system, and IFNLR1 mutations are closely related to hereditary hearing loss (ADN SHL). The zebrafish model construction and phenotypic analysis in this study were performed on the Shanghai Southern Model Bio- zebrafish platform. The researchers used a combination of genetic linkage analysis and whole exome sequencing to identify mutations in IFNLR1 that are responsible for autosomal dominant hearing impairment, and found that IFNLR1 c.296G> A (p.Arg99His) is likely A mutation in the causative gene of hereditary hearing impairment in this family. Figure 1. Identification of a mutation in IFNLR1 causing ADNSHL. Next, the researchers further evaluated the expression of the IFNLR1 gene in mice and mapped the expression pattern of Ifnlr1 in the inner ear of mice 30 days after birth (Fig 2A). In the cochlea, Ifnlr1 is expressed in the organ of Corti, Raisner's membrane, basement membrane, spiral ligament, sulcus cells, inner column cells and outer column cells (Fig 2B). Ifnlr1 is highly expressed in the inner hair cells (IHCs) and outer hair cells (OHCs) cilia, colocalized with the phalloidin (the epidermal plate of the outer hair cells of the cochlear basement membrane) (Fig 2G). Ifnlr1 was co-localized in cochlear nerve fibers and spiral ganglion neurons using a neurofilament heavy chain antibody (NF200) that specifically labels nerve fibers and neurons (Fig 2D, E). Ifnlr1 is highly expressed in the cytoplasm and cytoplasm of most cells of Curti; unlike IHCs, Ifnlr1 is not expressed in the nucleus of OHCs (Fig 2C, HJ). It was also observed that Ifnlr1 and phalloidin were co-localized in the vestibular epithelium (Fig 2F). Figure 2. Immunofluorescence analysis of ifnlr1 expression in the inner ear of P30 mice. The ifnlr1 knockdown zebrafish model was obtained using the zebrafish Morpholino (MO) Knockdown technique (Fig 3A, B). There was no significant change in the early development of zebrafish (from 32hpf to 4dpf) after ifnlr1 knocked down. In the control and ifnlr1-e4i4-MO injected fish, the circulation in the interstitial vessels (ISV) appeared normal. Knockdown of ifnlr1 does not induce organ-specific apoptosis or macrophage migration, suggesting that apoptosis and inflammation may not be responsible for the decrease in the number of hair cells. In the later stages of development, the ifnlr1 knockdown caused an abnormality in the zebrafish sputum inflation, and the intestinal lumen area expanded (Fig 3C-F). (The fish gills and human lungs are homologous organs, and abnormal fish gills indicate that the ifnlr1 gene mutation may also affect human lung function clinically.) The number of nerve mounds of ifnlr1 knocking down zebrafish is reduced compared to normal controls (Fig 3 I, M) . Using four markers, GFP (hair cells), SOX2 (supporting cells), BrdU (proliferating cells) and DAPI (nucleus), count the hair cells and support of each neural mound in 6dpf Tg (Brn3c:mGFP) transgenic zebrafish cell. Ifnlr1 knocked down the zebrafish juvenile fish, the hair cells and supporting cells in the lateral nucleus were lower than the wild type (WT) (Fig3O), and the number of BrdU-positive cells in the mutant was lower than that of WT (Fig3O, R), indicating hair cells and support. Loss of cells is associated with decreased cell proliferation. Figure 3. Phenotypes of ifnlr1 zebrafish morphants. MO-induced abnormalities were rescued by co-injecting zebrafish ifnlr1 mRNA with ifnlr1 MO into single-cell stage embryos. Rescue experiments showed that wild-type ifnlr1 mRNA can largely rescue the abnormal phenotype of fish aphid induced by ifnlr1-e4i4-MO, and the reduction of zebrafish hair cells and head nerve nucleus at 6dpf. These results suggest that IFNLR1 mutations cause hearing loss in the JS4842 family (Fig. 4). Figure 4. Coinjection of ifnlr1 mrna from non-mutant zebrafish rescued the phenotypes of non-inflated SB, hair cell loss and head neuromast loss (6 dpf) induced by ifnlr1-e4i4-MO. It was found by realtime-PCR that the cytokine receptor family member b4 (crfb4, also called interleukin (IL) 10 receptor 2, il10r2), jak1, tyrosine kinase 2 (tyk2), stat3, stat5, these Jak The gene associated with the /STAT signaling pathway was up-regulated after ifnlr1 knockdown (Fig. 5). Thus, it is speculated that the ifnlr1 mutation may contribute to hearing loss through the Janus kinase (Jak) 1 /signaling and transcriptional activator (sTAT) 3 signaling pathway. Figure 5. Jak1/Stat3 signalling pathway were upregulated in ifnlr1-morphant zebrafish. Pathological Analysis Equipments Pathological Analysis Equipments,Fsh Menopause Test Kit,Home Use Hcg Pregnancy,Hcg Rapid Test Car Test Strip Changchun ZYF science and technology CO.,LTD , https://www.zyf-medical.com
Hereditary hearing loss is highly genetically heterogeneous, and hundreds of gene mutations encoding multiple proteins may be associated with it. Patients with autosomal dominant non-syndromic hearing loss (ADNSHL) account for about 20% of hereditary hearing loss, and they usually develop from the initial high-frequency hearing loss to sublingual progressive hearing impairment. To date, 59 ADNSHL loci have been mapped and 36 pathogenic genes have been identified.
Interferon (IFN) lambda receptor 1 (IFNLR1, MIM 607404) belongs to the class II cytokine receptor family and is responsible for the recognition of cytokines and IFNs in the extracellular environment, as well as the initiation of intracellular signaling cascades leading to a series of responses. For example, hematopoiesis, immune system regulation, and cell growth and development. However, the function of IFNLR1 in the auditory system has not been described. 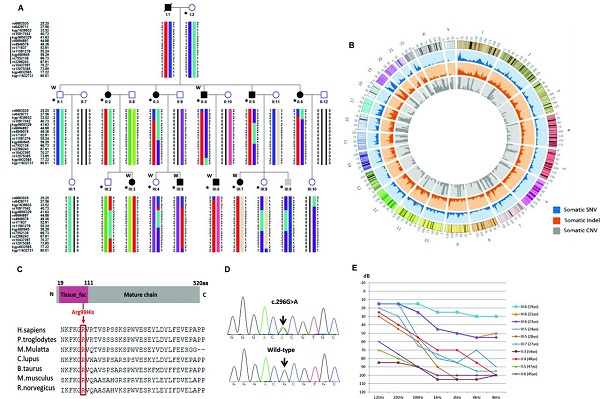
Using the zebrafish model to reveal for the first time that interferon lambda receptor mutation leads to clinical hereditary deafness
