Cloud order related product recommendation: m6A RNA whole transcriptome methylation sequencing m5C RNA whole transcriptome methylation sequencing m1A RNA whole transcriptome methylation sequencing Me-DIP sequencing Colorimetric detection of overall methylation levels Ultra-microRNA methylation sequencing RIP RNA Pull Down More than 10 points of m6A RNA methylation sequencing article----Cloud order bio-assisting No, Da Niu tells you how to study lncRNA methylation RNA methylation enters the ultra-micro era Xiaobai must see! Summary of methods for identification of overall level of RNA methylation RNA methylation in 20 points, coexistence of heat and strength Cloud Sequence Bio's latest m6A "RNA methylation" research summary - non-coding RNA articles Cloud Sequence Bio's Latest "RNA Methylation" Research Summary - Arabidopsis Cloud Sequence Bio's latest m6A "RNA methylation" research summary - virus articles Plant Cell: Arabidopsis found a new m6A RNA demethylation modifying enzyme Cloud order customer 12 points top article, teach you how to use RIP sequencing to play the molecular mechanism! 2018 Nature Magazine's Breakthrough Research Results--m5C RNA Methylation Cloud Sequence Creature Nature Breakthrough Research - RNA Methylation Newly Modified m1A Cloud Sequence Creature Hepatology: m6A RNA methylase METTL3 promotes the development of liver cancer 5 Inch Biometric Tablet,Sim Card Fingerprint Tablet,Wireless 4G Tablet With Barcode Scanner,Fingerprint Scanner Rugged Tablet Shenzhen Bio Technology Co., Ltd , https://www.hfsecuritytech.com
The authors studied mouse genomic methylation using a combination of SMRT-ChIP, RNA-seq and 6mA-IP seq. The study found that there is another DNA methylation modification in mouse embryonic stem cells - 6mA methylation; Alkbh1 is a demethylating enzyme modified by adenine methylation, adenine in the cells of Alkbh1 gene deletion mutant mice Increased levels of methylation lead to transcriptional silencing. It is revealed that 6mA methylation, an important component of mammalian apparent modification, plays a role in silencing gene expression in mammals, rather than activating gene expression in other organisms. 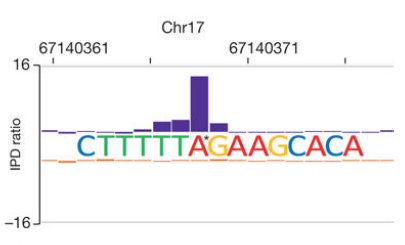
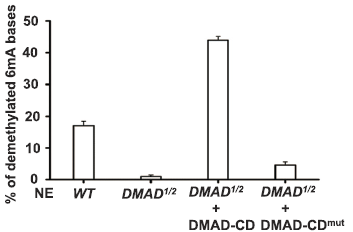
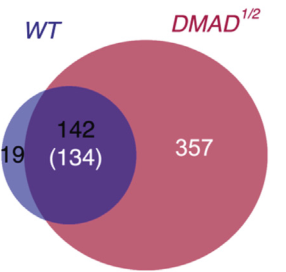
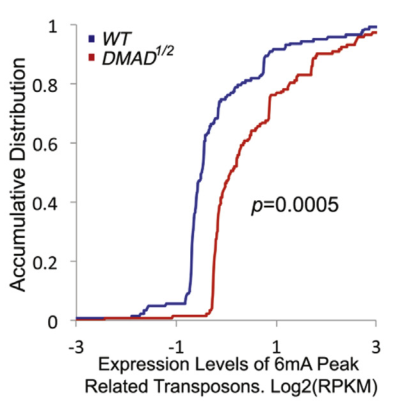
Using Drosophila embryonic stem cells as materials, the authors studied the 6mA modification in his genome and found that 6mA modification is widespread in Drosophila genome. This modification is regulated by endogenous demethylase DMAD and is dynamic during embryonic development. Variety. Further studies found that DMAD enzyme is an essential enzyme for Drosophila embryo development, which acts on the 6 mA site of the transposon. After demethylation, the expression of the transposon is inhibited. This article found a new type of DNA -6mA in Drosophila, and initially explored the regulatory mechanism of 6mA methylation. 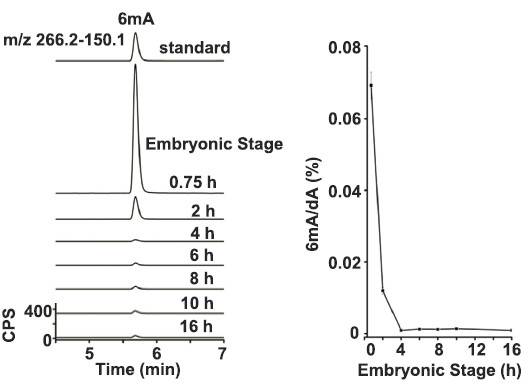
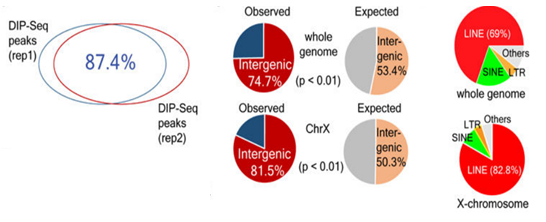
The article used three 6mA research techniques: 6mA-IP-Seq, 6mA-CLIP-Exo, and 6mA-RE-Seq to study the 6mA modification in the Drosophila genome and Chlamydomonas genome. It was found that 84% of the genes in the Drosophila genome are stably present with a 6 mA modification. And 6 mA is mainly distributed in the TSS region in the form of bimodal, which is related to the distribution of nucleosomes and is periodically distributed. Further binding to RNA-seq technology revealed that 6 mA is closely related to gene expression. The 6mA methylation modification in the genome of Chlamydomonas has new features. The 6mA modification is mainly distributed in the transcription initiation site and is distributed in bimodal form. Its main sequence features are C6mATG and G6mATC. Then, by analyzing the high-expression gene and low-expression gene detected by RNA-seq, it was found that the 6 mA concentration near the TSS with high expression gene was higher, while the 6 mA concentration near the TSS with low expression gene was lower, and 5 mC. The distribution characteristics are reversed. These data demonstrate that 6mA modification in eukaryotes can regulate gene expression. 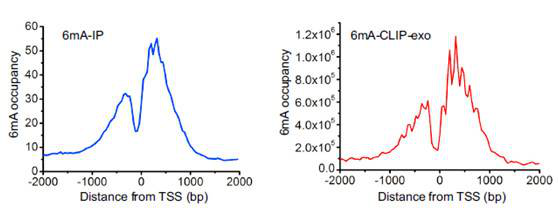
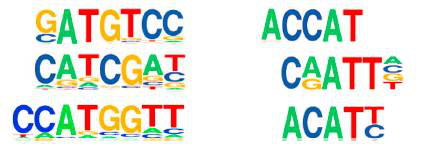
6mA distribution characteristics motif analysis 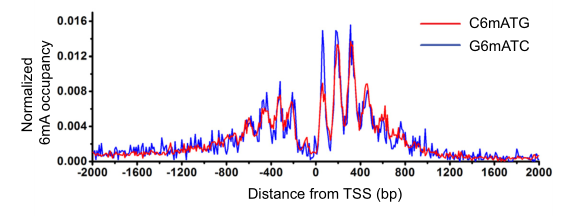
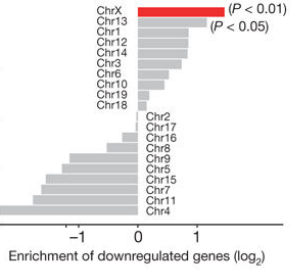
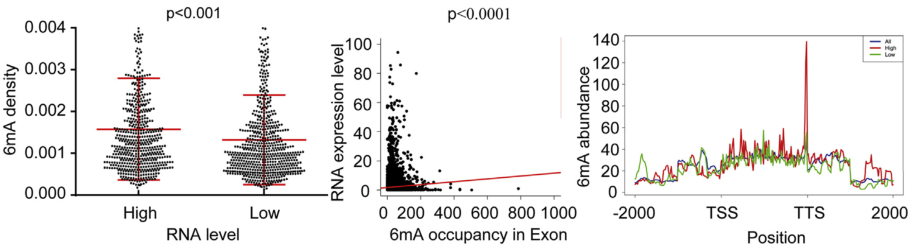
RNA-6mA has been shown to have important effects on gene expression in previous studies, but the research on the role of DNA-6mA in gene expression is not well understood. The author of this article first used the 6mA-IP-seq technique to analyze the DNA-6mA modification map: the exon region of the high expression level gene has a high 6mA abundance and is positively correlated with RNA expression level; 6mA The modification site is significantly enriched in G-protein coupled receptor (GPCR)-related genes, and it is predicted that 6mA methylation may play an important role in the regulation of GPCR-related gene expression. Subsequently, the regulation mechanism of DNA-6mA methylation was demonstrated by experiments such as knockout and overexpression of 6mA-IP-qPCR, ALKBH1 gene and N6AMT1 gene. In summary, the significance of this article is to decipher the human DNA N6-methyladenine (6mA) modification map for the first time, revealing the regulatory mechanism and biological function of DNA-6mA methylation modification in the human genome. 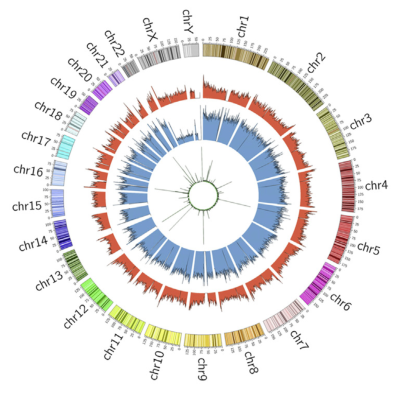
Cloud Sequence Bio has pioneered the development of DNA-6mA sequencing technology. The principle is similar to MeDIP's co-immunoprecipitation sequencing technology. The 6mA fragment is enriched by specific antibodies, amplified and subjected to high-throughput sequencing and methylation analysis, and the whole genome DNA is quickly and efficiently drawn with a small amount of data. A map of methylation levels.
Customers only need to provide genomic DNA for the relevant samples, and Cloud Sequence Bio completes the entire service flow from 6mA-IP enrichment, library preparation, on-machine sequencing to data analysis.
Optimized experimental process:
The enrichment efficiency of 6mA-IP is the key to determining data quality. The Cloud Sequence Bio 6mA-IP experiment uses pre-validated commercial antibodies and well-optimized experimental procedures with high efficiency and specificity.
Strict quality control:
The key steps of the experiment are strictly quality control, and the quality of the experiment is monitored throughout the process to ensure that customers receive quality data.
Professional bioinformatics analysis:
Cloud Sequence Bio has a strong bioinformatics team that can meet the needs of customers for in-depth data analysis.
2, sample requirements
Sample type: DNA sample without degradation and no RNA contamination (for all species with reference genomes).
Sample demand: no less than 10ug
Sample concentration: not less than 50ng/ul
Sample transportation and storage:
Sample transport: The sample was placed in a 1.5 mL Eppendorf tube, the parafilm was sealed, transported on dry ice, and the DNA was transported in ice packs.
Sample storage: cell sample or fresh tissue block diced, liquid nitrogen stored at -80 ° C after cryopreservation; DNA samples can be -20 ° C in the short term, to avoid repeated freezing and thawing.
3, data analysis
Standard analysis:
(1) Identification of DNA methylation enrichment peaks
Through high-throughput sequencing and bioinformatics analysis, identify the methylated enriched genomic region, the default p <= 1e-5 (specific parameters based on the report, the cloud sequence organism will adjust the parameters according to the data) Significant methylation zone.
Note: Make an Input for each sample set to remove the genomic background and reduce the false positive rate. 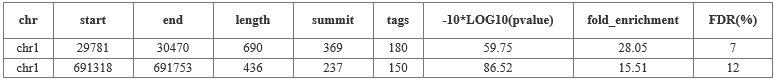
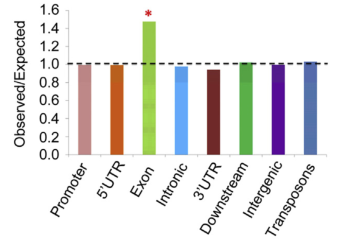
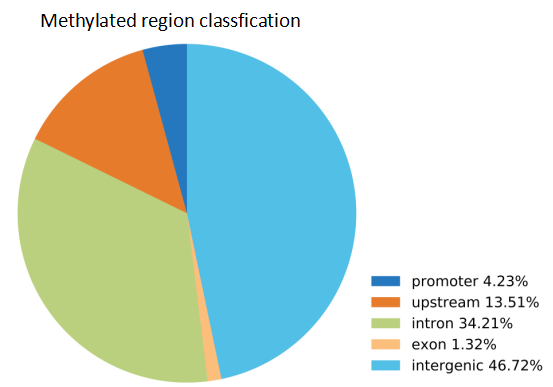
After the enrichment peak is identified, a pile of genomic location information is obtained, and the enriched peak is annotated by the nearest neighbor gene through bioinformatics analysis, and the enriched peak is used as a promoter according to the position of the peak midpoint relative to the known gene. Peak, upstream peak, intron peak, exon peak, intergenic peak. 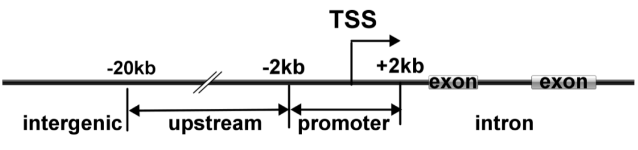
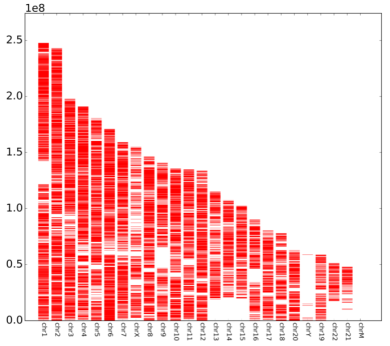
Cloud-sequence organisms use diffReps software to identify differentially methylated regions (DMRs). The default p-value <0.0001, fold change >=2 is the threshold for the differential methylation zone. 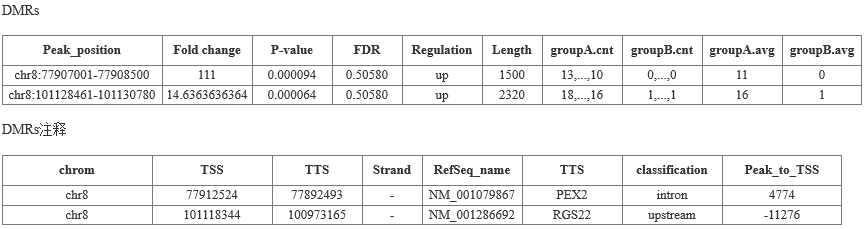
OBJECTIVE: To functionally classify differentially methylated genes and to identify functional entries that are significantly enriched. 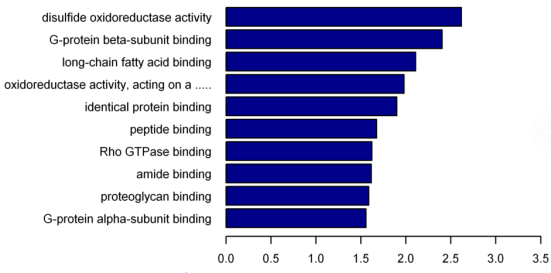
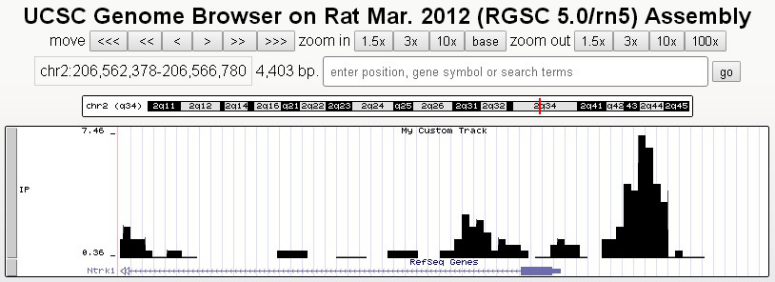
(1) DNA methylation site motif analysis
The DNA methylation modification preference sequence can be determined by sequence analysis combined with bioinformatics to find the DNA methylation feature Motif. 
Based on the annotation information, a consensus and specific methylation modification site or gene between the groups is drawn. 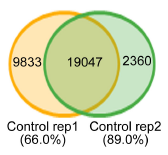
DNA-6mA methylation sequencing
Shanghai Yunxu Biological Technology Co., Ltd.
Shanghai Cloud-seq Biotech Co., Ltd.
Address: 3rd Floor, Building 20, No. 518, Zhangzhu Road, Songjiang District, Shanghai 
Telephone Fax Website:
Email: .c
See how the apparently modified -6mA methylation helps IF soar!
DNA methylation modification is one of the hotspots of epigenetic research. We usually think that DNA methylation is cytosine methylation (5-methylcytosine, 5mC), but I don't know that with the rapid development of sequencing technology, researchers A new DNA methylation modification, DNA-6mA methylation, has been discovered in eukaryotes (Drosophila, fungi, Chlamydomonas reinhardtii, C. elegans, etc.), and DNA-6mA methylation can affect Gene expression plays a key regulatory role in the growth and development of plants and animals, and is closely related to a variety of human diseases including cancer. These findings have successfully set off a research boom in DNA-6mA, making DNA-6mA a hot topic in the field of epigenetics. Next, I will introduce DNA-6mA in several classic cases.
DNA methylation on N6-adenine in mammalian embryonic stem cells.NATURE, IF: 44.958
6mA analysis of the relationship between 6mA and gene expression
6mA-IP seq analysis
N6-Methyladenine DNA Modification in Drosophila.Cell, IF: 31.398
6mA distribution mA in vitro demethylation analysis
Combined analysis of 6mA-IP seq and RNA-seq
N6-Methyldeoxyadenosine Marks Active Transcription Start Sites in Chlamydomonas. Cell, IF: 31.398
Peak analysis
N6-Methyladenine DNA Modification in the Human Genome. Molecular Cell, IF: 14.248
DNA-6mA correlation analysis
Cloud Sequence Bio - 6mA-IP -seq One-stop Service
1. Technical service
One-stop service:
(2) Notes on enrichment peaks in DNA methylation regions
(3) Distribution of DNA methylation-enriched peak regions in genes
(4) Identification of differential DNA methylation regions (DMRs) and annotations
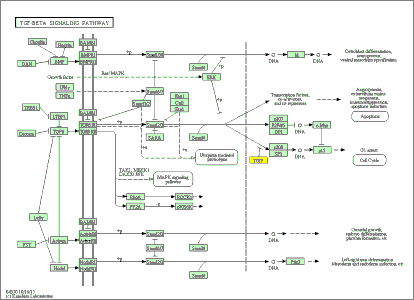
(5) Analysis of GO (Gene Ontology) and Signal Pathway in Differential DNA Methylation Region
(6) DNA methylation visualization
Advanced analysis:
(2) DNA methylation site sharing and specificity analysis
A wonderful review in the past:
